

GenoDive v. 3.0 now available!
GenoDive is the premier Mac-application for population genetic analyses. The program was made with both diploids and polyploids in mind: all analyses can be used with multiple ploidy levels, for some analyses even up to hexadecaploids! GenoDive features the intuitive user interface that you expect on a Mac, as well as a large number of different types of analyses.
To learn more about GenoDive, click here to download the manual.
Many different types of inferences
 GGenoDive features many different types of statistical inferences, some of which are not available in any other population genetics software. Included inferences are among others: a flexible Analysis of Molecular Variance, many different estimators of population differentiation, AMOVA-based K-means clustering, assigning genotypic identity (clones) to individuals, testing for clonal reproduction, Hardy-Weinberg equilibrium, calculation of the hybrid index for individuals, 23 ways of calculating genetic distances, assignment tests, different types of Mantel tests, and both PCA and PCoA.
GGenoDive features many different types of statistical inferences, some of which are not available in any other population genetics software. Included inferences are among others: a flexible Analysis of Molecular Variance, many different estimators of population differentiation, AMOVA-based K-means clustering, assigning genotypic identity (clones) to individuals, testing for clonal reproduction, Hardy-Weinberg equilibrium, calculation of the hybrid index for individuals, 23 ways of calculating genetic distances, assignment tests, different types of Mantel tests, and both PCA and PCoA. Analyse multiple datasets... simultaneously!
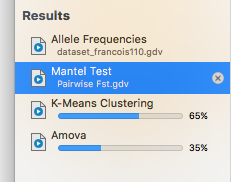 All Macs that are sold nowadays contain multiple processor cores. However, most programs are able to use only a part of the power of these new computers as they employ only a single processor at a time. GenoDive automatically detects the number of processors present in the computer and divides lengthy tasks over them all. This makes analyses in GenoDive much faster than in other programs. Calculations always take place in the background, so you can perform other analyses while waiting for the results of a more lengthy analysis.
All Macs that are sold nowadays contain multiple processor cores. However, most programs are able to use only a part of the power of these new computers as they employ only a single processor at a time. GenoDive automatically detects the number of processors present in the computer and divides lengthy tasks over them all. This makes analyses in GenoDive much faster than in other programs. Calculations always take place in the background, so you can perform other analyses while waiting for the results of a more lengthy analysis. Multiple data formats
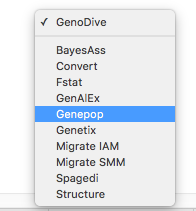 GenoDive can handle three different types of data: markerdata (e.g. SNPs from RAD sequencing), distance matrices (e.g. pairwise genetic distances between populations), and generic other data (e.g. the coordinates of populations), which can be combined in the statistical inferences. Importing data into GenoDive is easy: just open a file and GenoDive will automatically determine what type of data it is and will load the data into memory. GenoDive has support for many different file formats including Fstat, Genepop, Spagedi, GenAlEx, BayesAss, Structure, and Migrate, saving you time spent on tediously reformatting your input files.
GenoDive can handle three different types of data: markerdata (e.g. SNPs from RAD sequencing), distance matrices (e.g. pairwise genetic distances between populations), and generic other data (e.g. the coordinates of populations), which can be combined in the statistical inferences. Importing data into GenoDive is easy: just open a file and GenoDive will automatically determine what type of data it is and will load the data into memory. GenoDive has support for many different file formats including Fstat, Genepop, Spagedi, GenAlEx, BayesAss, Structure, and Migrate, saving you time spent on tediously reformatting your input files. Easily include and exclude observations
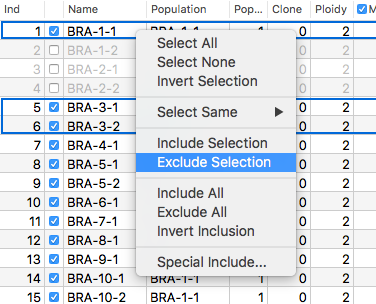 Most real-life datasets are far from perfect, some loci may be more reliable than others and there will always be individuals whose DNA-extraction yielded low quality DNA. In most programs assessing their influence means quitting the program, editing the inputfile and then restarting the program again with the new file. GenoDive makes it easy to include or exclude loci, individuals, clones, ploidy levels, populations, or even whole groups of populations, so the effect of those outliers on the outcome of the analyses can be assessed.
Most real-life datasets are far from perfect, some loci may be more reliable than others and there will always be individuals whose DNA-extraction yielded low quality DNA. In most programs assessing their influence means quitting the program, editing the inputfile and then restarting the program again with the new file. GenoDive makes it easy to include or exclude loci, individuals, clones, ploidy levels, populations, or even whole groups of populations, so the effect of those outliers on the outcome of the analyses can be assessed. A clear view over your data
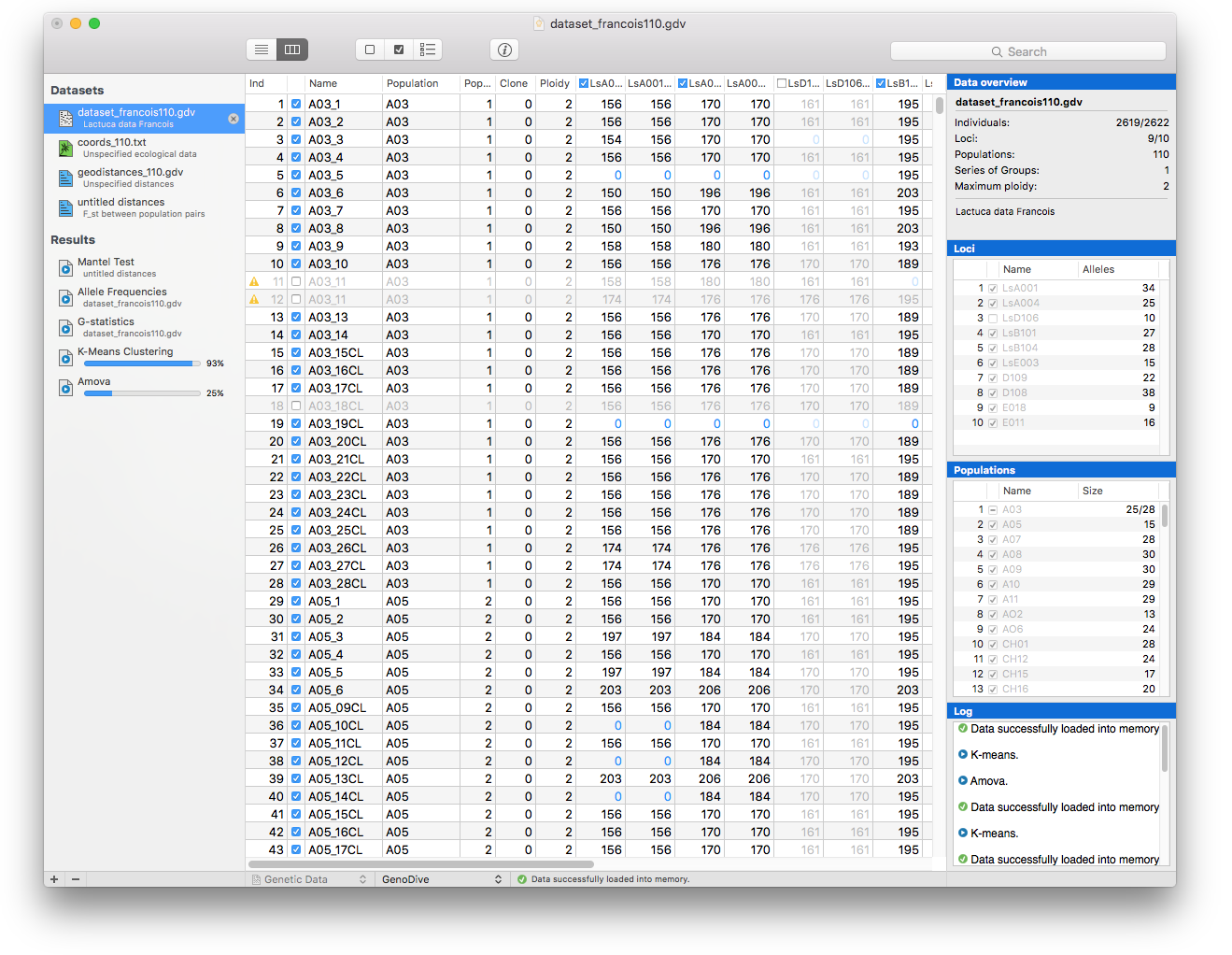 GenoDive has a clear, uncluttered, interface that makes it easier for you to keep an overview of your data and analyses. You can choose between viewing your data as plain text or viewing it as a matrix. The Inspector panel shows the most important information for the current data: how many individuals are included per population, the number of populations groups, which loci are included, the number of alleles per locus and a log with the analyses that have been performed on this dataset.
GenoDive has a clear, uncluttered, interface that makes it easier for you to keep an overview of your data and analyses. You can choose between viewing your data as plain text or viewing it as a matrix. The Inspector panel shows the most important information for the current data: how many individuals are included per population, the number of populations groups, which loci are included, the number of alleles per locus and a log with the analyses that have been performed on this dataset.


